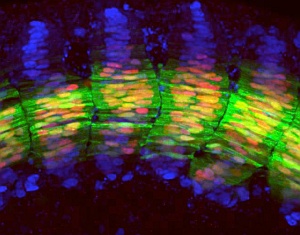
Confocal fluorescence microscopy image of the trunk region of a transgenic zebrafish embryo, showing arrays of muscle cell nuclei (blue) and expression of Engrailed (red) and Eng2a proteins.
Progenitor cells are stem cells with a predetermined destiny. During embryonic development, they differentiate into their target cell type upon stimulation by signaling molecules, such as those of the ‘Hedgehog’ and bone morphogenic protein (BMP) pathways. Cells often respond to multiple signals, but the way in which progenitor cells deal with two opposing signals, such as Hedgehog and BMP, has remained unclear. Philip Ingham at the A*Star Institute of Molecular and Cell Biology and colleagues have now shed new light on how progenitor cells respond to multiple signals by elucidating the molecular interactions underlying muscle cell development in zebrafish.
The researchers studied the molecular events involved in the specification of two specialized types of muscle fiber found in the zebrafish embryo, both express the gene engrailed2a (eng2a) and are known to respond to Hedgehog signaling molecules. “The zebrafish is easy to breed and to manipulate genetically, making it an ideal model species for studying the genetic control of cell specification in vertebrate embryos,” explains Ingham.
Combining sophisticated transgenic approaches with the exquisite confocal-based imaging for which the zebrafish embryo is renowned (see image), Ingham’s team identified a minimal regulatory element upstream of eng2a that is required to limit its expression to the specialized muscles. Surprisingly, however, they found that this element lacks consensus-binding sites for the transcription factors Gli1 and Gli2a that mediate Hedgehog signaling.
“We were puzzled by this finding because Hedgehog signaling activity is known to play a crucial role in the specification of these muscles in the zebrafish embryo,” says Ingham. Using a novel tissue-culture-based assay, the researchers screened for other potential regulators of the eng2a regulatory element and identified Smad proteins—transcription factors that mediate BMP signaling—as strong repressors.
Returning to the zebrafish embryo, they found strong accumulation of activated Smad proteins in the nuclei of muscle progenitor cells, but not in those that express eng2a. Moreover, by experimentally blocking the nuclear accumulation of activated Smad proteins, the researchers showed that they could elicit eng2a expression even when Hedgehog signaling was compromised. Most strikingly, they found that maximal Hedgehog signaling caused depletion of nuclear Smad proteins, suggesting that ‘crosstalk’ between the two pathways occurs at the posttranslational level. They also obtained evidence that this crosstalk may be mediated by direct interaction between the Gli and Smad proteins.
“Our results suggest a novel mechanism for crosstalk between the Hedgehog and BMP pathways that may be deployed in a variety of contexts,” concludes Ingham.
The A*STAR-affiliated researchers contributing to this research are from the Institute of Molecular and Cell Biology.



