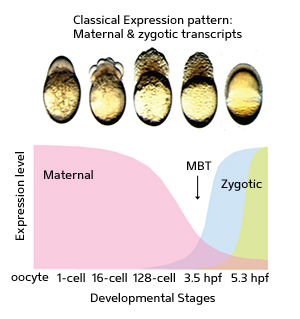
A chart of messenger RNA expression levels at different stages of zebrafish development. Analysis of transcripts at each stage indicated the presence of three major ’superclusters’, one expressed purely from maternal mRNA (purple), followed by a subset of maternal ’pre-MBT’ mRNA transcripts (blue) that usher in the onset of the MBT, at which point the embryo exclusively produces its own zygotic mRNA population (green).
© 2011 CSH Press
Offspring make their first move towards independence surprisingly early in life. After fertilization, they initiate protein production — a process that is guided exclusively by maternal messenger RNA (mRNA) molecules stored within the egg. Several hours later, however, the embryo’s own genes begin to switch on and produce zygotic mRNA, a threshold known as the mid-blastula transition (MBT).
In order to clarify the poorly understood mechanisms underlying MBT, Sinnakaruppan Mathavan at the A*STAR Genome Institute of Singapore and co-workers have set out to assemble a detailed catalogue of the mRNA molecules present at different developmental stages in zebrafish. Zebrafish are ideal models for such studies, as their embryos develop entirely outside of the mother’s body, mature rapidly and can be cultivated in large quantities. “Our aim was to produce sequencing data that can be used as a genomic resource for the zebrafish community,” explains Mathavan.
The researchers used high-throughput sequencing technology to collectively analyze embryonic mRNA content at multiple developmental stages, from the unfertilized egg to the post-MBT embryo. Based on the sequencing results, they identified three ‘superclusters’ of genes with peak expression at maternal, pre-MBT or zygotic phases (see image). Many of these genes could be further classified within smaller clusters with similar expression profiles. Their analysis also revealed numerous previously uncharacterized mRNA transcripts and alternative versions of known mRNAs.
In order for mRNAs to be efficiently translated into proteins, they must undergo a process known as polyadenylation, in which each molecule gets ‘tailed’ with a long stretch of adenine nucleotides. However, Mathavan and co-workers noted that many early mRNAs appeared to be unmodified and thus dormant. “Until now, it was believed that all maternal RNAs are polyadenylated and translationally active soon after fertilization, at the one-cell stage,” says Mathavan. “We discovered that this is not true.”
In fact, the researchers identified two subsets of maternal mRNAs. The first is polyadenylated from the beginning, and these appear to drive the earliest stages of postfertilization embryogenesis. The other group is initially inactive, but undergoes gradual adenylation as a prelude to MBT, at which point these transcripts give rise to proteins that facilitate MBT onset and the production of zygotic mRNA.
The study provides fresh insights into the different stages of zebrafish development. Mathavan notes that similarly illuminating studies could soon be performed using mice. “It would be more challenging to obtain early-stage embryos,” he says. “However, new high-throughput sequencing platforms require fewer starting materials. It should therefore be possible to perform similar studies with mammals in the near future.”
The A*STAR-affiliated researchers contributing to this research are from the Genome Institute of Singapore.



