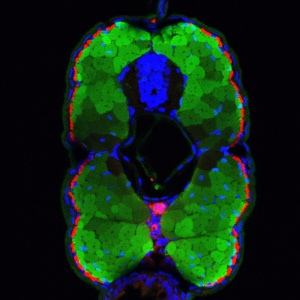
Cross section through a 6-day-old transgenic zebrafish larva showing the fast twitch specific expression of GFP under the control of the endogenous Sox6 promoter (transcriptional) and 3’UTR sequences (translational); the slow twitch fibers are outlined in red; the nuclei are stained blue.
Understanding how cells maintain their determined state once specified during embryogenesis is a major aim of developmental biology. Philip Ingham at the A*STAR Institute of Molecular and Cell Biology and co-workers have uncovered a gene regulatory network underlying the stability of skeletal muscle fiber-type differentiation.
During early development, cells derived from a tissue layer called mesoderm develop into muscle progenitor cells, which subsequently differentiate into two distinct muscle-fiber types: slow-twitch fibers, which contract slowly and work for a long time without getting tired; and fast-twitch fibers, which contract quickly but use more energy and tire quickly.
The transcriptional regulator Sox6 plays an important role in fast-twitch fiber specification. In the zebrafish embryo, transcription of Sox6 is specifically repressed in slow-twitch progenitors by the transcription factor Prdm1a, the gene for which is in turn activated by the signalling molecule Sonic hedgehog (Shh) secreted from the notochord, the flexible rod-shaped precursor of the inter-vertebral discs found in all vertebrate embryos. Notably, the expression of Prdm1a is only transient, raising the question as to how Sox6 repression is maintained in slow-twitch fibers post-embryonically.
“We took advantage of the high resolution imaging afforded by the zebrafish to analyze the spatial regulation of Sox6 expression,” says Ingham.
By generating transgenic zebrafish lines, the researchers identified regulatory sequences upstream of the Sox6 gene that play a crucial role in determining whether muscle progenitors differentiate into fast- or slow-twitch muscle fibers. They showed that this regulatory element specifically drives Prdm1a-dependent Sox6 expression in fast-twitch progenitors. However, the same element became transcriptionally active in the slow-twitch fibers of larval stages. Further experiments revealed that the endogenous Sox6 gene is also activated in larval slow-twitch fibers, whereas the Sox6 protein remains restricted to fast-twitch fibers.
“This surprising finding suggested to us that the expression of Sox6 is regulated partly at the level of translation rather than just transcriptionally as expected,” says Ingham.
The researchers next showed that Prdm1a induces the specific expression in slow-twitch progenitors of a microRNA molecule encoded by a non-coding region of another gene. They found that this microRNA, called miR-499, in turn repressed the translation of Sox6 in these cells.
“This ‘double-hit’ mechanism involving both transcriptional and translational repression, sets up a feedback loop that ensures Sox6 is not expressed even after Prdm1a protein dissipates, thereby maintaining as well as initiating the slow-twitch muscle lineage,” says Ingham. “Similar mechanisms are likely to control the specification of other cell types.”
The A*STAR-affiliated researchers contributing to this research are from the Institute of Molecular and Cell Biology.



